Portfolio
Professional portfolio for Sean Maden. Includes programming projects, utilities, dashboards, and research projects.
lute
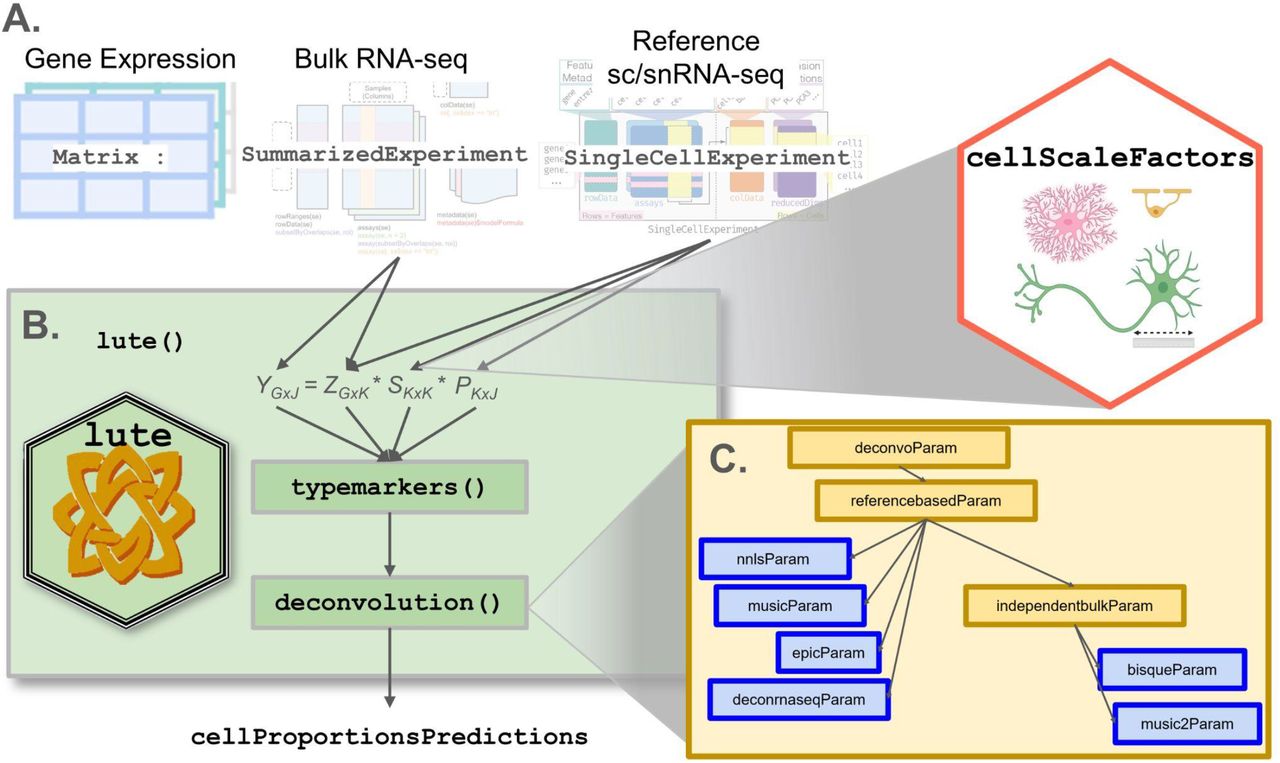
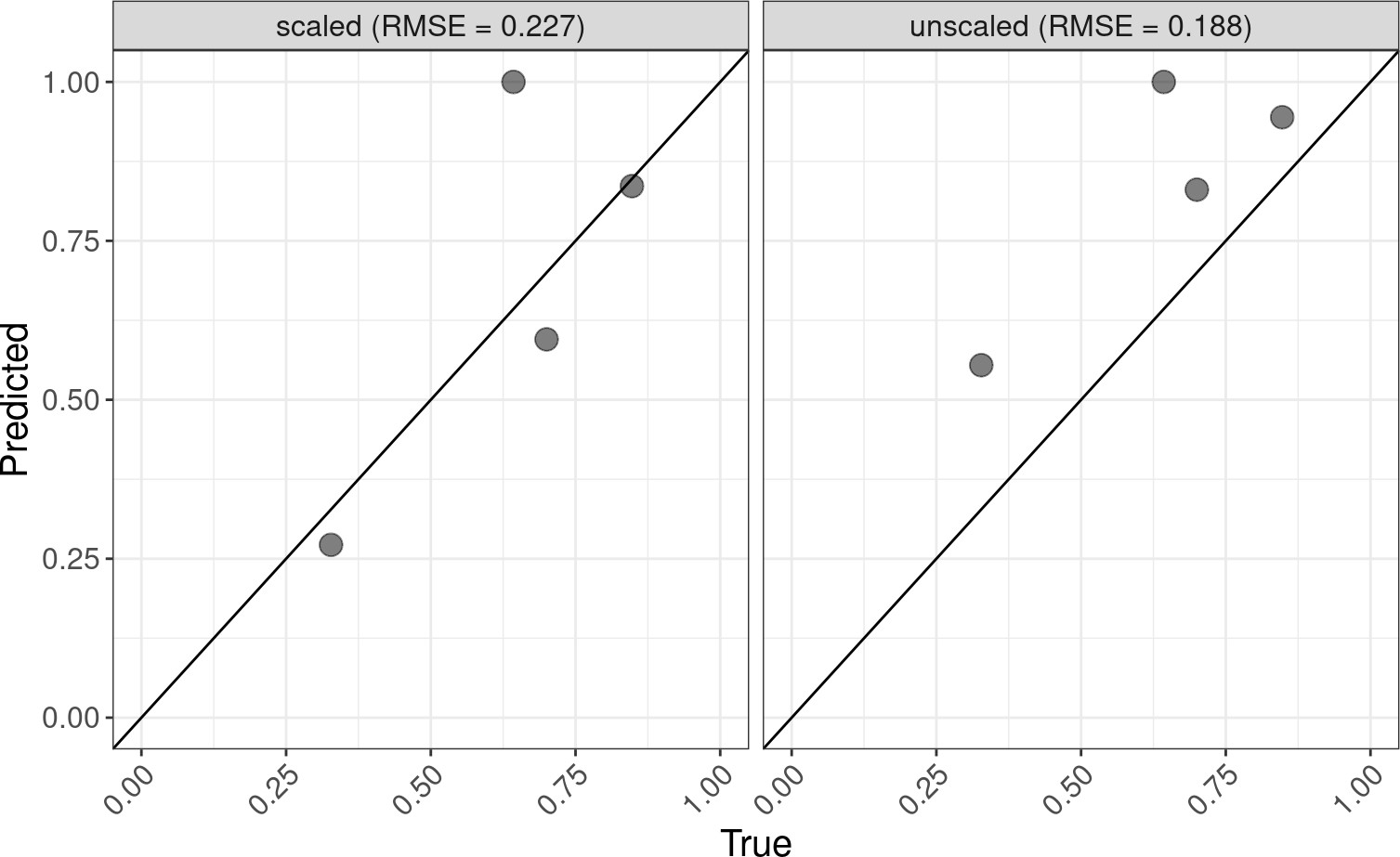
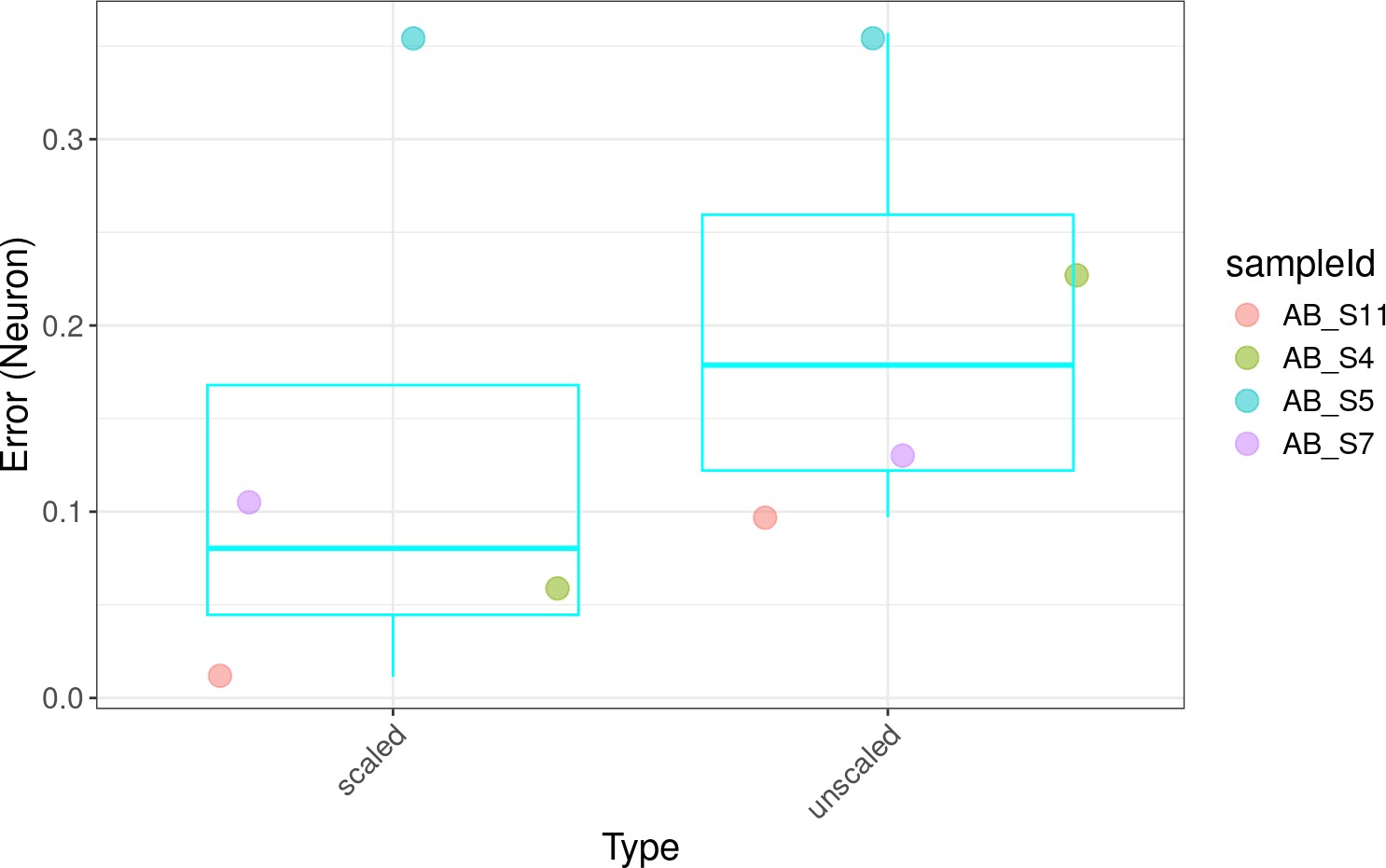
2023, R/Bioconductor software packageThe lute R package supports bulk transcriptomics deconvolution experiments. It gets its name from the word "deconvolute," the problem of predicting cell amounts from cell mixtures. Users can use the lute() function to select for markers and perform deconvolution either with or without a cell size scale factor normalization (see ?lute for details). For normalizations, data may be incorporated from the cellScaleFactor package and passed to the s argument for transformation of cell reference expression data prior to deconvolution.

recountmethylation
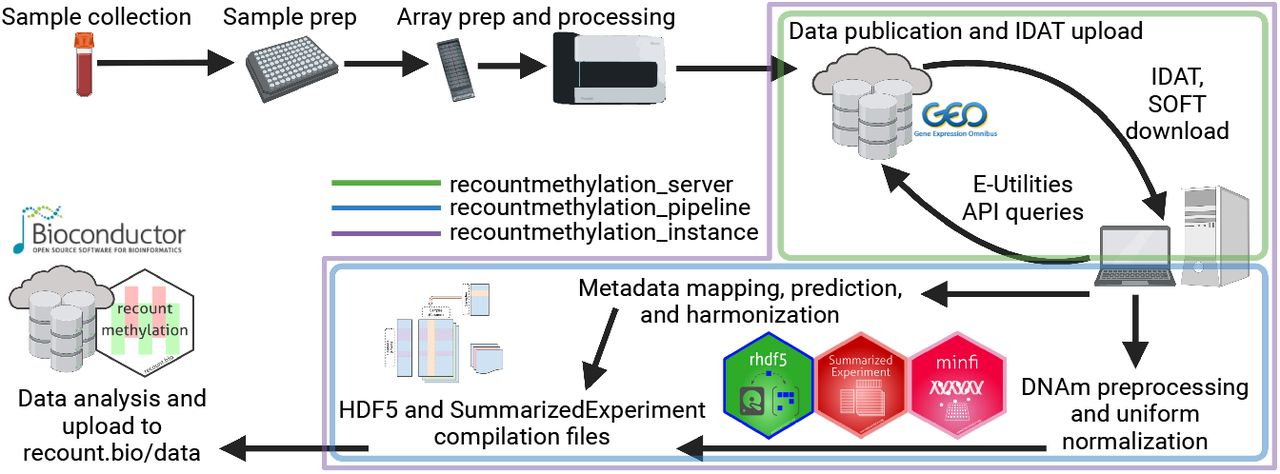
R/Bioconductor software packageResources for cross-study analyses of public DNAm array data from NCBI GEO repo, produced using Illumina's Infinium HumanMethylation450K (HM450K) and MethylationEPIC (EPIC) platforms. Provided functions enable download, summary, and filtering of large compilation files. Vignettes detail background about file formats, example analyses, and more. Note the disclaimer on package load and consult the main manuscripts for further info.

cgmappeR
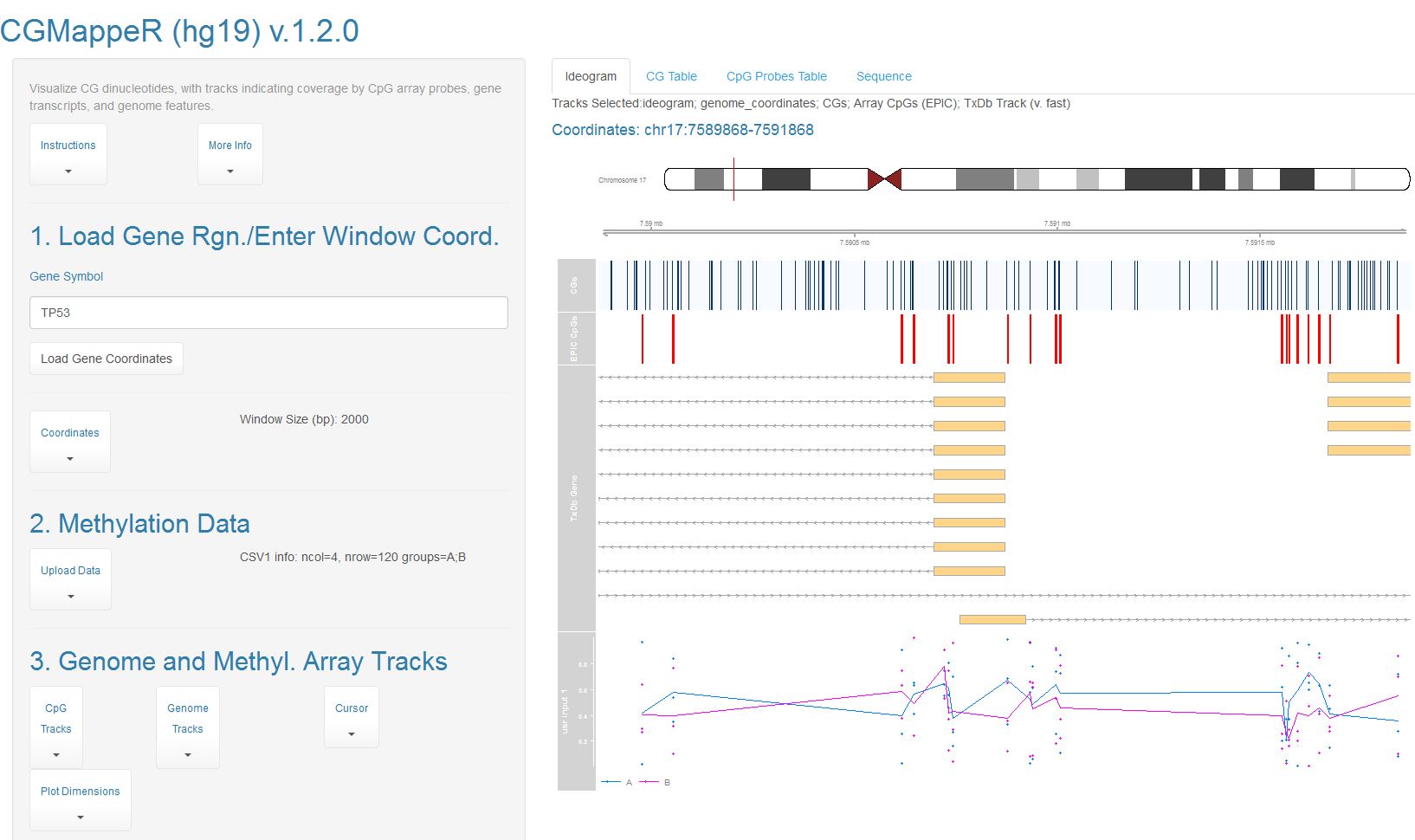
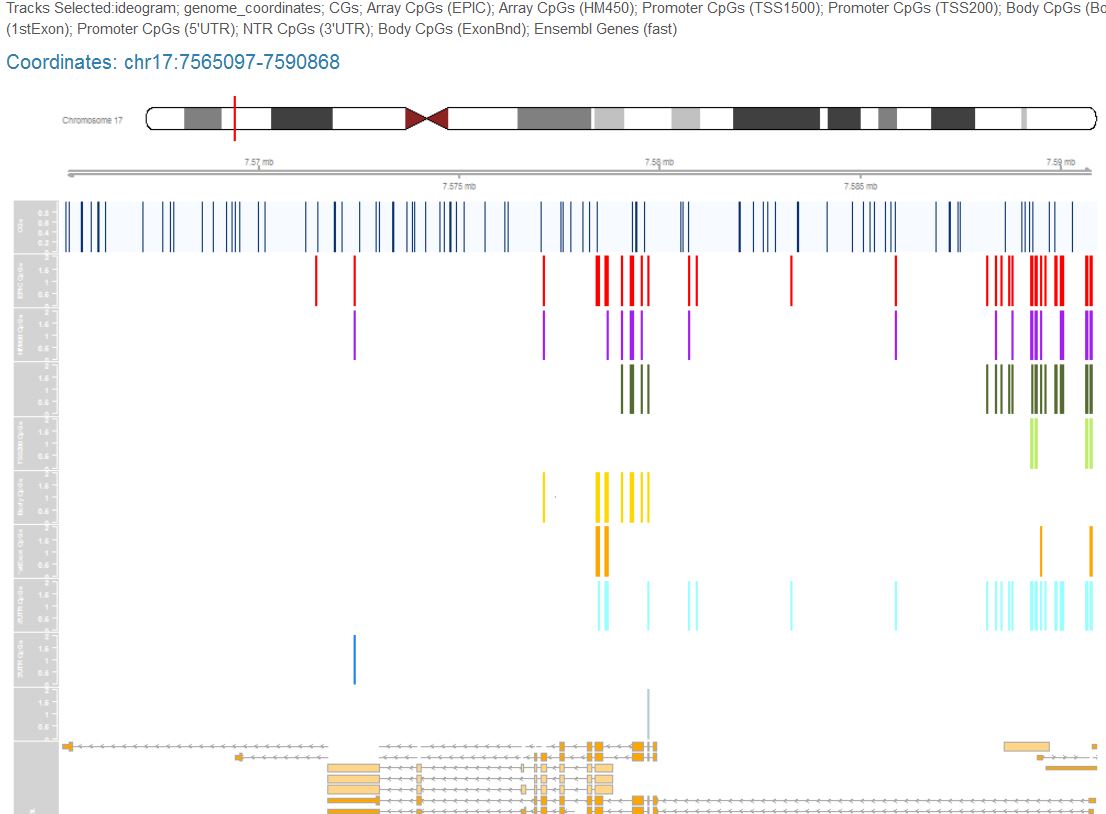
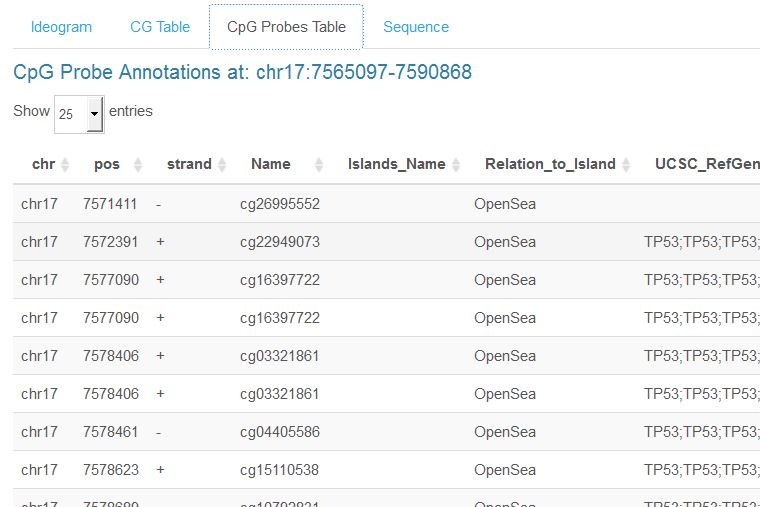
R/Shiny dashboardAn R/Shiny dashboard to map and visualize cytosine-guanine (CG/CpG) dinucleotides and microarray-targeted probes. Map and visualize genomic CpGs, plot your own input DNA methylation microarry data, query genes and gene-proximal regions, generate high-quality ideograms, and export CpG sequence, CpG coordinates, and probe annotations from custom genome views.
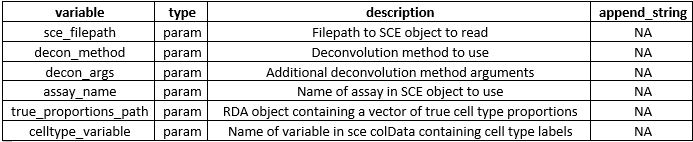
r-nf_deconvolution
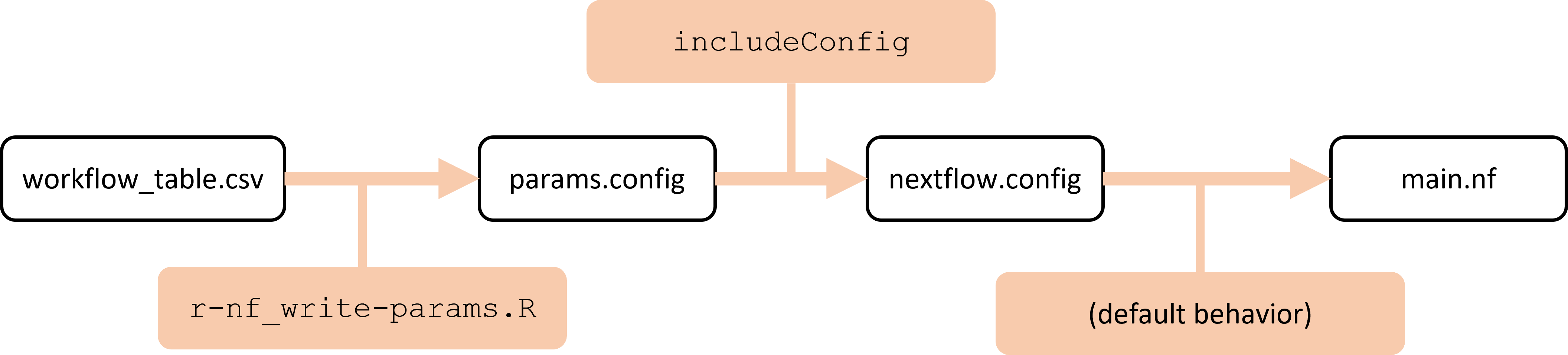
Nextflow workflowDeconvolution NextFlow workflow using R and Rscripts. Scripts provided in the sh and yml folders show which dependencies are required and how to install them. For most runs, you will at minimum need a recent version of NextFlow, R, and the nnls, SummarizedExperiment, and SingleCellExperiment libraries, with additional required dependencies for other deconvolution methods besides the non-negative least squares (NNLS) method. You can use the provided .yml file to set up a conda virtual environment.


geoarraydigest (GAD)
Python appletResource for summarizing and tracking new array samples published to NCBI's Gene Expression Omnibus (GEO) database. Geo Array Digest (GAD) resource is an automated messaging service that monitors available of public array datasets made available in GEO database. The GAD service was constructed by scheduling queries to the GEO API using the Entrez Programming Utilities software. Automated status updates to the GAD Bot Twitter account are made using the twitteR R package in a snakemake workflow. It is maintained by Sean Maden, PhD (GitHub: metamaden, Twitter: @MadenSean).

Generated by Copilot AI, version 2025, Microsoft.
© 2025 Sean Maden. All rights reserved.